What Kind Of Mutation Occurs When One Base Is Changed To Another At A Single Location In The Dna?
Mutations aren't simply grouped co-ordinate to where they occur — oftentimes, they are besides categorized by the length of the nucleotide sequences they impact. Changes to short stretches of nucleotides are called gene-level mutations, considering these mutations affect the specific genes that provide instructions for various functional molecules, including proteins. Changes in these molecules can have an affect on whatsoever number of an organism'south physical characteristics. Equally opposed to gene-level mutations, mutations that alter longer stretches of DNA (ranging from multiple genes upward to entire chromosomes) are called chromosomal mutations. These mutations oftentimes accept serious consequences for afflicted organisms. Because gene-level mutations are more than common than chromosomal mutations, the following sections focus on these smaller alterations to the normal genetic sequence.
Base substitution
Base substitutions are the simplest type of cistron-level mutation, and they involve the swapping of one nucleotide for another during Deoxyribonucleic acid replication. For example, during replication, a thymine nucleotide might exist inserted in identify of a guanine nucleotide. With base exchange mutations, only a unmarried nucleotide within a factor sequence is changed, so but one codon is affected (Figure ane).
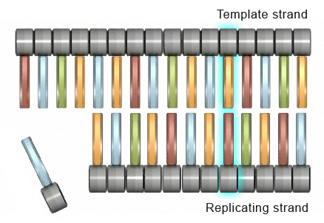
Figure one: Merely a unmarried codon in the factor sequence is changed in base substitution mutation.
Although a base exchange alters only a single codon in a factor, it tin still accept a pregnant impact on protein production. In fact, depending on the nature of the codon change, base of operations substitutions tin lead to iii different subcategories of mutations. The start of these subcategories consists of missense mutations, in which the altered codon leads to insertion of an incorrect amino acrid into a protein molecule during translation; the 2d consists of nonsense mutations, in which the altered codon prematurely terminates synthesis of a protein molecule; and the third consists of silent mutations, in which the altered codon codes for the same amino acrid equally the unaltered codon.
Insertions and deletions
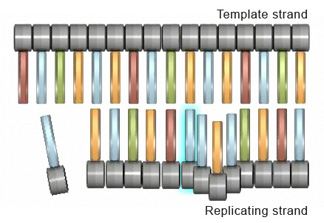
Figure 2: During an insertion mutation, the replicating strand \"slips\" or forms a wrinkle, which causes the actress nucleotide to exist incorporated.
Insertions and deletions are 2 other types of mutations that can touch cells at the gene level. An insertion mutation occurs when an extra nucleotide is added to the DNA strand during replication. This can happen when the replicating strand "slips," or wrinkles, which allows the extra nucleotide to exist incorporated (Figure 2). Strand slippage tin also lead to deletion mutations. A deletion mutation occurs when a wrinkle forms on the Deoxyribonucleic acid template strand and subsequently causes a nucleotide to be omitted from the replicated strand (Effigy iii).
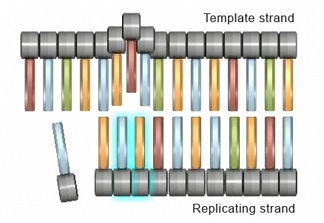
Effigy 3: In a deletion mutation, a contraction forms on the Dna template strand, which causes a nucleotide to be omitted from the replicated strand.
Frameshift mutations
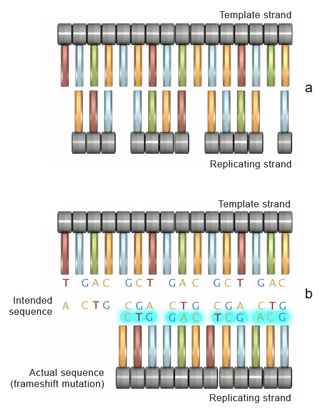
Figure 4: If the number of bases removed or inserted from a segment of DNA is not a multiple of three (a), a dissimilar sequence with a different fix of reading frames is transcribed to mRNA (b).
Insertion or deletion of ane or more than nucleotides during replication can too lead to another type of mutation known as a frameshift mutation. The outcome of a frameshift mutation is consummate amending of the amino acrid sequence of a protein. This alteration occurs during translation because ribosomes read the mRNA strand in terms of codons, or groups of 3 nucleotides. These groups are called the reading frame. Thus, if the number of bases removed from or inserted into a segment of DNA is not a multiple of 3 (Effigy 4a), the reading frame transcribed to the mRNA will be completely changed (Figure 4b). Consequently, once it encounters the mutation, the ribosome will read the mRNA sequence differently, which can upshot in the production of an entirely different sequence of amino acids in the growing polypeptide concatenation.
To better understand frameshift mutations, let'south consider the analogy of words as codons, and letters within those words as nucleotides. Each word itself has a divide meaning, as each codons represents one amino acid. The following sentence is composed entirely of three-letter words, each representing a three-letter codon:
THE Large BAD FLY HAD ONE Red EYE AND I BLU Middle.
Now, suppose that a mutation eliminates the sixth nucleotide, in this instance the letter "G". This deletion means that the letters shift, and the remainder of the judgement contains entirely new "words":
THE BIB ADF LYH ADO NER EDE YEA NDO Beak LUE YE.
This error changes the relationship of all nucleotides to each codon, and finer changes every single codon in the sequence. Consequently, at that place is a widespread modify in the amino acid sequence of the protein. Lets consider an example with an RNA sequence that codes for a sequence of amino acids:
AUG AAA CUU CGC AGG AUG AUG AUG
With the triplet code, the sequence shown in figure 5 corresponds to a protein fabricated of the following amino acids:
Methionine-Lysine-Leucine-Arginine-Arginine-Methionine-Methionine-Methionin
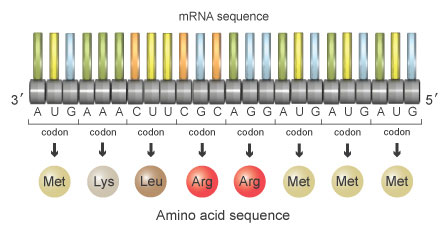
Figure 5: This sequence of mRNA codes for the amino acids methionine-lysine-leucine-arginine-arginine-methionine-methionine-methionine.
At present, suppose that a mutation occurs during replication, and it results in deletion of the fourth nucleotide in the sequence. When separated into triplet codons, the nucleotide sequence would at present read as follows (Effigy 6):
AUG AAC UUC GCA GGA UGA UGA UG
This series of codons would encode the following sequence of amino acids:
Methionine-Asparagine-Phenylalanine-Alanine-Glycine-Stop-STOP
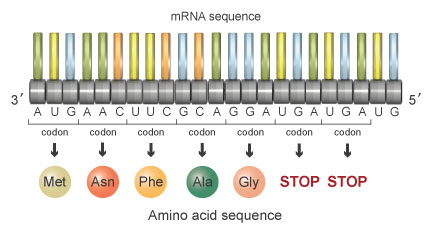
Figure 6: If the 4th nucleotide in the sequence is deleted, the reading frame shifts and the amino acid sequence changes to methionine-asparagine-phenylalanine-alanine-glycine-Finish-STOP.
Each of the cease codons tells the ribosome to stop protein synthesis at that betoken. Consequently, the mutant protein is entirely unlike due to the deletion of the fourth nucleotide, and it is also shorter due to the advent of a premature stop codon. This mutant poly peptide will be unable to perform its necessary function in the cell.
Source: http://www.nature.com/scitable/topicpage/dna-is-constantly-changing-through-the-process-6524898
Posted by: martinhignisfat.blogspot.com


0 Response to "What Kind Of Mutation Occurs When One Base Is Changed To Another At A Single Location In The Dna?"
Post a Comment